The Homeobox and its related genes have played an
interesting but challenging role in developmental biology and now in cancer
pathways. The genes related to this 180 base pair section of DNA are the genes
which control the development or organs and the time at which these development
occur. Furthermore the structure of this gene collection is preserved across an
dramatically large number of species, the human included. Thus it was
interesting to see a paper in NEJM discussing the mutation of a specific Homeobox
gene, HOX B 13, as relates to prostate cancer.
The novel HOXB13
G84E variant is associated
with a significantly increased risk of hereditary prostate cancer. Although the
variant accounts for a small fraction of all prostate cancers, this finding has
implications for prostate-cancer risk assessment and may provide new
mechanistic insights into this common cancer.
Now this appears as a significant new finding and we would
like to examine this a bit. The HOX genes are quite unique in their
functioning. They are built about a core Homeobox segment, which is preserved
across chromosomes and species, and is hen connected with variable regions on
differing chromosomes to generate some 4X13 possible genes (HOX (A,B,C,D) (1…13)).
These genes are core to the morphological and embryological development of a
broad range of species.
Now HOX B 13 is one of many Homeobox based genes. These
genes are distributed across 4 chromosomes and have a fixed part called the
homeobox part and a variable part. In a sense it is similar to the fixed and variable regions we see in the immune system. The gene is created as below:

Homeobox genes are clustered in the
chromosomes and are expressed in the body in the same order in which they occur
in the chromosomal DNA. The HOX genes, the concatenation of the respective Homeobox and its
variable part are named by chromosome location as A,
B, C, D, and then by number 1 through 13 at present. The number reflects what
makes the Homeobox genes of interest, namely the genes
control the development of the embryos, namely they control what cells do as a
part of the development of an entity. The process goes from head to tail, and
the numbering goes from the earliest or anterior to the latest or posterior
elements in the development process. Thus HOX A 1
relates to an early development and HOX B 13
would refer to a later development of the embryo. The sequencing is shown below.
Retinoic acid activates the Homeobox genes sequentially in
development.
Now the Ewing study examined patients with specific changes:
Given the consistent
evidence of prostate-cancer linkage to 17q21-22 markers in our multiplex families
with hereditary prostate cancer, we designed a targeted sequencing strategy to
analyze 2009 exons of 202 genes contained in the most likely genomic interval
defined by our fine-mapping studies. … Probands from four families were
observed to have the same nonsynonymous mutation in HOXB13,
a change of adenosine for
guanine (transition, c.251G→A) in the second position of codon 84
(GGA→GAA),
resulting in a nonconservative substitution of glutamic acid for glycine (G84E)
The question is perhaps where does the term Homeobox come
from. From Gehring and Hiromi we have the definition:
The term
"homeosis" (originally spelled "homoeosis") was proposed by
Bateson (8) to describe the transformation of one structure of the body into
the homologous structure of another body segment. Homeotic transformation can result,
for example, from abnormal regeneration of amputated structures (epigenetically)
or from germ-line mutations
Thus the Homeobox genes are key to the development of
embryos. They also lead to the discussions
Scott states:
Homeotic genes control
cell fates during the development of all animals, as was first revealed by studies
of the Drosophila homeotic gene complexes … Many of these genes contain a homeobox, a 180 bp sequence
of DNA which encodes an evolutionarily conserved DNA binding
domain, the homeodomain … A plethora of mammalian homeobox genes have been reported, among
which 38 are located in four clusters. A new nomenclature for the mammalian Hox
genes, approved … The new names take advantage of the elegant arrangement
of the genes to provide a logical nomenclature system
rather than the names given when the genes were discovered. The new system is initially
designed only for vertebrate genes, although it is to be hoped that
similar systems will be useful, and adopted, for other animals. In order to preserve
as much
clarity
in the literature as possible, it has been agreed by a large number of workers in the
field and by the nomenclature committees that homeobox genes not located
within the Hox complexes should not be given names containing the word 'Hox'.
There are four
clusters of Hox genes … now to be known as A, B, C, and D. Based on sequence similarity the
genes can
be sorted into 13 'paralog' groups, each group having, in
most cases,
a representative
in each complex. The order of paralogs along
the chromosome is preserved in the four complexes. The genes
within a
complex
are
transcribed in the same direction and are numbered according to their
paralog
group from 1 at the 3' end to 13 at the 5' end. In several
cases
a representative
of a paralog group is absent from a complex, in which case the corresponding gene number is
omitted …
HOX genes are key to the development of the embryo, it
creates the head to tail and sets up the control of the development of the
organs. As Lohmann and McGinnis report:
Hox genes
play a major role in the morphological diversification of the anteroposterior
body axis of animal embryos by switching the fates of segments between
alternative developmental pathways . In their role of controlling segment
diversity, Hox proteins are responsible for many different morphological structures
and cell types within a given segment. But it is still largely a mystery how a
single Hox gene
can determine a morphological trait at a specific location within a segment,
and why that trait does not appear elsewhere in the same segment or in other
segments.
… morphological and
transcriptional responses to Hox genes
can be highly local, sometimes only in a single cell, allowing
one Hox gene to control a
cavalcade of different traits within one segment and between different
segments, depending on the information present. Another important lesson that
we can learn from the papers of Rozowski and Akam and Brodu et al. is that, during development, Hox genes act at all levels in the
developmental hierarchy.
If they act very far down in the hierarchy, as in
these two cases, then the output is subtle, with Hox genes acting as cell-type switches rather than as major
developmental pathway switches. If they are acting (apparently) far up in the
hierarchy, then the fate switch is more dramatic, which is most beautifully
demonstrated in the famous four-winged fly. But even at this general level,
context is still crucial: loss of Ubx in
the haltere does not generate a leg, but a wing.
There are many debates still raging regarding Homeobox and
Robert presents an interesting report summarizing some of them. His paper is
worth the reading. It builds on the evo-devo issue, evolution and development,
the ontogeny recapitulates ontogeny. Namely if the same HOX genes are present across
many species, and preserved in structure, then is there really an underlying
commonality across species.
We provide the details on the various HOX genes below. They
all have the form as we had shown earlier and they are all numbered in a
sequence consistent with what we have shown earlier.
We now show from Kim et al the development of the pathway
for the HOX B 13 that we have been discussing. It inhibits CDK and that in turn
inhibits the activation via E2F of the cell cycle. It is the inhibition of the
cell cycle that is of the most concern.
As Kim et al demonstrate the HOX
B 13 blocks p21 and in turn CDK2 keeping the RB pathway from entering the cell
into cell cycle reproduction. They state:
Taken together, the
results of this study demonstrated the presence of a novel pathway that helps understand
androgen-independent survival of prostate cancer cells. These findings suggest
that upregulation of HOXB13 is associated with an additive growth advantage of
prostate cancer cells in the absence of or low androgen concentrations, by the
regulation of p21-mediated E2F signaling.
Now Ewing at al conclude as follows:
In summary, we have
used linkage analysis in combination with targeted massively parallel
sequencing to identify a recurrent mutation in HOXB13
that is associated with
early-onset and hereditary prostate cancer. From a clinical perspective, testing
for germline mutations in BRCA1/2 is recommended in some families, since mutations
in these breast-cancer–susceptibility genes are associated
with elevations in the risk of prostate cancer, particularly for BRCA2 However, neither of these genes has been
shown to contribute to hereditary prostate cancer. HOXB13
G84E is associated with a
significantly increased risk of hereditary prostate cancer.
This work suggests
that future DNA sequencing studies using next-generation technology and study populations
enriched for genetic influence (as evidenced by an early age at onset and
positive family history) may identify additional rare variants that will
contribute to familial clustering of prostate cancer. Although HOXB13
mutations will be identified
in a minority of men with prostate cancer, rare genetic lesions can identify
pathways that are found to be abnormal in more common, sporadic cases.
This leaves one to somewhat guess as to how prevalent this
mutation is. The rough numbers given in the Weing paper is about 1.5%. It also begs the question of why as a mutation which is apparently
inherited the progression of the cancer is so slow. Ewing at al show that the
odds ration can be as high as 32.5:1 when the mutation is present. The age at
diagnosis is lower with an odds ratio of 2:1 but with the problem one sees in
pathway control one wonders why the cancer does not appear much earlier as seen
in BRCA.
Thus this paper raises several questions:
1. The Homeobox mutation is a predisposing genetic risk factor.
If tested and found positive for the factor what should be done next.
Mastectomy is often what BRCA patients undergo, does this mean prophylactic
prostatectomy?
2. The pathway seems to be somewhat understood. The E2F
family control the pathway and HOX B 13 controls that pathway. It blocks it to
some degree. What can happen to HOX B 13 to cause this change in non-mutated
individuals.
3. Can the disease propensity be regulated by genetic
pathway control, is this possible as an alternative prophylactic measure.
4. What other pathway elements should be considered. Specifically, if we have a mutation on HOX B 13 then must we have other genes also altered to up surge cell replication. If so which ones. Is HOX B 13 merely a predisposing element. Also is there a HOX B 13 type change in other PCa?
5. Most importantly, why does it take so long for the cancer
to develop, are there precursor hits somewhere and this this just eliminates
other hits?
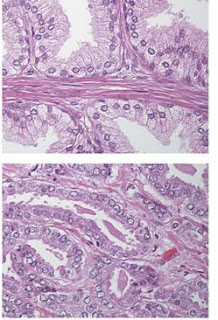
Ewing et al have an interesting slide showing normal versus
HOX B 13 prostate cells and we replicate it below from the paper.
In
the top slide we see well-structured prostate cells with basal and luminal
layers not showing and aberrant growth, no PIN. In the slide below from a HOX B
13 patient with a mutation of the form: GGA to GAA Glycine Glutamic acid (See
Ewing et al).
References:
-
Ewing, C., et al, Germline Mutations in HOX B 13
and Prostate Cancer Risk, NEJM, Jan 2012 V 366 N 2 pp 141-149.
- Jung, C., et al, HOX B 13 Homeodomain Protein
Suppresses the Growth of Prostate Cancer, Can Res 2004 V 64 pp 3046-3051.
- Kim Y, et al, HOX B 13 promotes Androgen
Independent Growth, Molecular Cancer, 2010 Vol 9-124.
-
Lohman, I., W. McGinnis, HOX Genes, Current
Biology, 2002, V 12 pp 514-516.
-
Robert, J., Interpreting the Homeobox; Metaphors
of Gene Action and Activation in Development and Evolution, Evo & Dev, 2001
V 3:4 pp 287-295.
-
Schwartz, J., Homeobox Genes, Fossils, and the
Origin of the Species, Anat Rec 1999 V 257 pp 15-31.
-
Scott, M., A Rational Nomenclature for
Vertebrate Homeobox, Nu Acid Res 1993 V 21 No 8 pp 1687-1688.
- Gehring, W., Y. Hiromi, Homeotic Genes and the Homeobox, Ann Rev Gen 1986 V 20 pp 147-173.