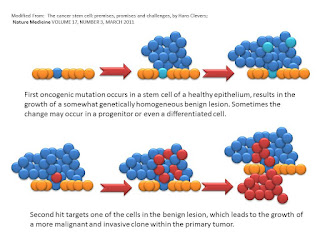
Now Clevers suggests a four step process, albeit with limited experimental evidence, but an superb start. It is as follows:
The above are the first two steps. Perhaps a dysplsia or neoplasia but with the kernel of a stem cell. This is the first "hit" theory. The epithelium starts to grow in a strange manner. Say a polyp in the colon or HGPIN in the prostate. Then we see a second hit and the formation of extraepithelial growth.
Then the third hit for the author and we see transmission via the blood stream. Then the fourth hit and the explosion from a few to almost all cancer stem cells.
Whether this is a good or bad model is yet to be seen. As Clevers states:
Central to the cancer stem cell (CSC) concept is the observation that not all cells in tumors are equal. The CSC concept postulates that, similar to the growth of normal proliferative tissues such as bone marrow, skin or intestinal epithelium, the growth of tumors is fueled by limited numbers of dedicated stem cells that are capable of self-renewal. The bulk of a tumor consists of rapidly proliferating cells as well as postmitotic, differentiated cells. As neither of these latter two classes of cells has the capacity to self-renew, the contribution of these non-CSC tumor cells to the long-term sustenance of the tumor is negligible.
The increased focus on the CSC is truly needed because if it is indeed a key paradigm in cancer then it and not large tumor masses should be examined. Clevers concludes with:
Epilogue: are CSCs and clonal evolution mutually exclusive?
To date, the CSC field has treated tumors as genetically homogeneous entities, by and large ignoring the fact that the observed tumor heterogeneity may result from underlying genetic differences. However, it is well known that most solid tumors show extensive genomic instability. Moreover, genetic defects in a large variety of molecules that are involved in the maintenance of the integrity of the genome are well-known drivers of oncogenesis. Even in a disease like CML, so clearly driven by stem cells, clonal evolution can be seen at work when imatinib is administered: the malignancy becomes tumor-resistant through the emergence of clones that carry mutations in the target of imatinib, the BCR-ABL1 fusion gene75. And the progression of CML into ALL blast crisis is caused by the emergence of subclones that harbor inactivating lesions in the cyclin-dependent kinase inhibitor 2A (CDKN2A, also known as ARF) gene in addition to the BCR-ABL1 translocation76. The evidence for clonal evolution in the pathogenesis of cancer is so overwhelming that it appears inescapable that all models should be integrated with it.
The recent rapid advances in DNA sequencing are now allowing the global analysis of genomic changes of cancer cells. These analyses have confirmed many previously known common genetic alterations in cancer, and they have also revealed some new common mutations as well as unexpectedly large numbers of rare mutations. As a next step, this technology can be applied to chart genetic heterogeneity within individual tumors as well as between primary tumors and their local recurrences and metastases. It should thus be possible to map, in both space and time, the genetic evolution of a tumor.
The last sentence is the most compelling. Cancer may be more than just a cellular disease, it may require the spatial domain as well. This is an exceptionally good review and should be a focus for future research.